Abstract
Background
Children with Down syndrome (DS), which caused by an extra copy of chromosome 21, have a 20-fold increased risk for acute lymphoblastic leukemia (ALL) compared with non-DS children. Recent genome-wide studies have revealed that DS-ALL have uncommon genetic alterations such as mutations in JAK2 , NRAS , and KRAS , mutations or overexpression of CRLF2 , suggesting the unique biological features of DS-ALL distinct from non-DS ALL. However, most of previous studies regarding molecular analysis of DS-ALL have been limited to genomic and transcriptomic characterizations, and no study showed comprehensive genetic/epigenetic overview in DS-ALL.
Materials and Methods
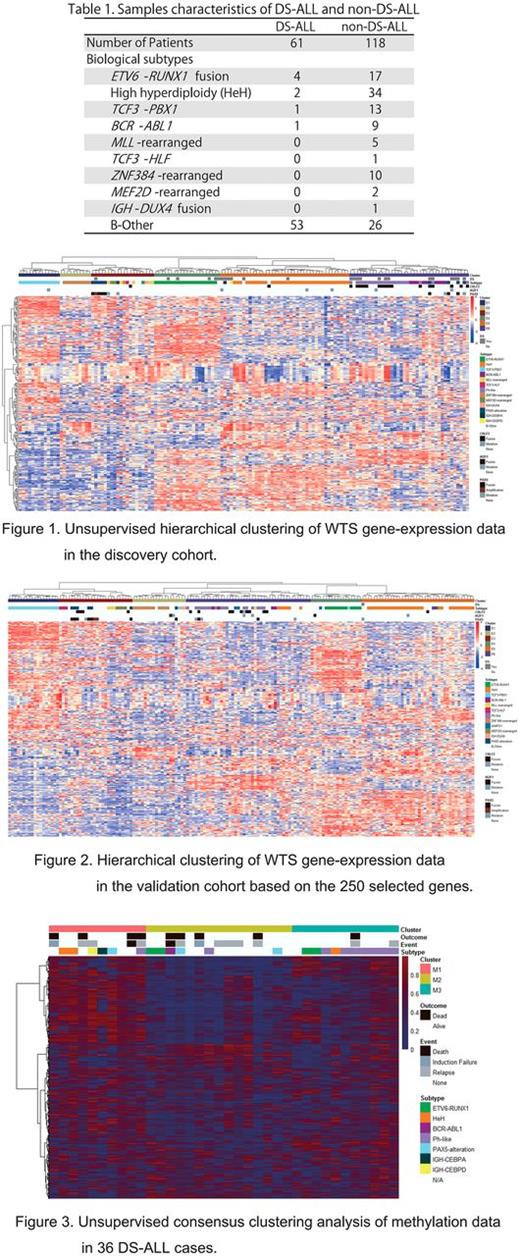
Our cohort included 179 pediatric B-cell precursor ALL samples (61 DS-ALL and 118 non-DS-ALL). Biological subtypes are detailed in Table 1. We performed targeted deep sequencing for seven ALL-related genes in 61 DS-ALL, SNP array analysis in 43 DS-ALL, and whole transcriptome sequencing (WTS) in 25 DS-ALL and 118 non-DS-ALL samples. WTS data from 153 ALL in TARGET database were also used in this study as a validation cohort. Additionally, methylation array analysis was performed in 36 DS-ALL, 17 non-DS-ALL, and 20 DS patients without ALL. To validate gene mutations detected by WTS, Sanger sequencing was also conducted. To cluster gene expression profiling, we applied the hierarchical clustering method for expression profiling and consensus clustering for methylation profiling.
Results
Targeted deep sequencing revealed frequent gene mutations of JAK2 (23%) and RAS (31%) pathway. SNP array analysis elucidated several recurrent chromosomal deletions affecting actionable gene loci; IKZF1 (33%), PAX5 (30%), EBF1 (15%), CDKN2A (34%), and RB1 (7%). Gains of whole chromosome X (18%) and 8 (10%) were also recurrently identified. Expression profiling stratified 143 samples into 6 clusters (E1-E6 clusters), which were well correlated with biological subtypes of ALL (Figure 1). Intriguingly, DS-ALL cases were clustered into four clusters, suggesting the heterogeneity of DS-ALL. Cluster E3 included one DS-ALL, which was clustered to the branch of PAX5 -rearranged cases. SNP array analysis revealed this case had focal amplification of PAX5 . All DS-ALL cases with ETV6 - RUNX1 fusion were classified into cluster E4, which also included all ETV6 - RUNX1 fusion cases of non-DS-ALL. This cluster included one DS-ALL case without ETV6 - RUNX1 fusion. This particular case had a homozygous deletion of ETV6, implicating ETV6 - RUNX1 -like signature. DS-ALL with high hyperdiploidy (HeH) were classified into cluster E5, which also included most of HeH samples of non-DS-ALL. Among other 5 DS-ALL cases in this cluster, 2 cases showed IGH - CEBPA fusion and IGH - CEBPD fusion, respectively. Cluster E6 was characterized by BCR - ABL1 fusion and Ph-like expression profiles. In our cohort, we identified 7 DS-ALL cases as Ph-like signature, which included all JAK2 mutated samples. In this cluster, PAX5 mutation and IKZF1 mutation were detected in 2 DS-ALL cases, respectively. Notably, by using 250 genes in our hierarchical clustering, TARGET database samples were divided to similar 6 clusters (Figure 2). On the other hand, Methylation array analysis revealed unique hypermethylation of promoter regions of RUNX1 in DS-ALL. Methylation of this regions were confirmed in B-cell of DS without ALL, but not in non-DS-ALL. Furthermore, methylation array analysis categorized DS-ALL into 3 clusters (Figure 3). Cluster M1 included IGH - CEBPA fusion and IGH - CEBPD fusion cases. This cluster had worse prognosis than other 2 clusters.
Conclusion
Our integrated analysis enlightened the genetic and epigenetic heterogeneity of DS-ALL. Importantly, DS-ALL had genetic features corresponding to each biological subtypes same as non-DS-ALL. Our analyses also elucidated that DS-ALL had unique subtypes such as IGH - CEBPA fusion, IGH - CEBPD fusion, or PAX5 alterations. It may be a reason for worse prognoses of DS-ALL. Our result indicated risk classification or therapy of DS-ALL should be tailored by each biological subtype.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal